Scientists demonstrated an explicit and unique mode of polymerase fidelity modulation
Typically not assisted by proofreading, the RNA-dependent RNA polymerases (RdRPs) encoded by the RNA viruses may need to independently control its fidelity to fulfill virus viability and fitness. However, the precise mechanism by which the RdRP maintains its optimal fidelity level remains largely elusive.
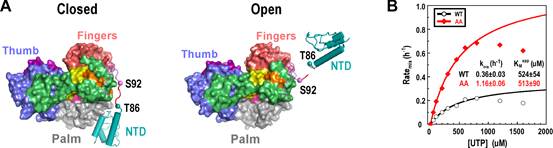
By solving 2.1–2.5 A resolution crystal structures of the classical swine fever virus (CSFV) NS5B, an RdRP with a unique naturally fused N-terminal domain (NTD), the research group led by Prof. GONG Peng in Wuhan Institute of Virology of the Chinese Academy of Sciences identified high-resolution intra-molecular interactions between the NTD and the RdRP palm domain.
In order to dissect possible regulatory functions of NTD, the scientists designed mutations at residues Y471 and E472 to perturb key interactions at the NTD–RdRP interface. When crystallized, some of these NS5B interface mutants maintained the interface, while the others adopted an ‘open’ conformation that no longer retained the intra-molecular interactions. Data from multiple in vitro RdRP assays indicated that the perturbation of the NTD–RdRP interactions clearly reduced the fidelity level of the RNA synthesis, while the processivity of the NS5B elongation complex was not affected.
Collectively, their work demonstrates an explicit and unique mode of polymerase fidelity modulation and provides a vivid example of co-evolution in multi-domain enzymes.
The results have been published in Nucleic Acids Research entitled "A unique intra-molecular fidelity-modulating mechanism identified in a viral RNA-dependent RNA polymerase".
This work was supported by the National Key Research and Development Program of China, National Natural Science Foundation of China, Chinese Academy of Science, and Ministry of Science and Technology of China.